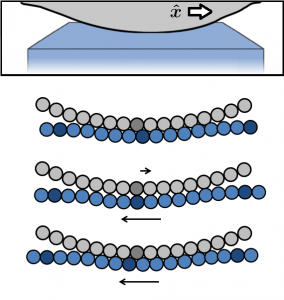
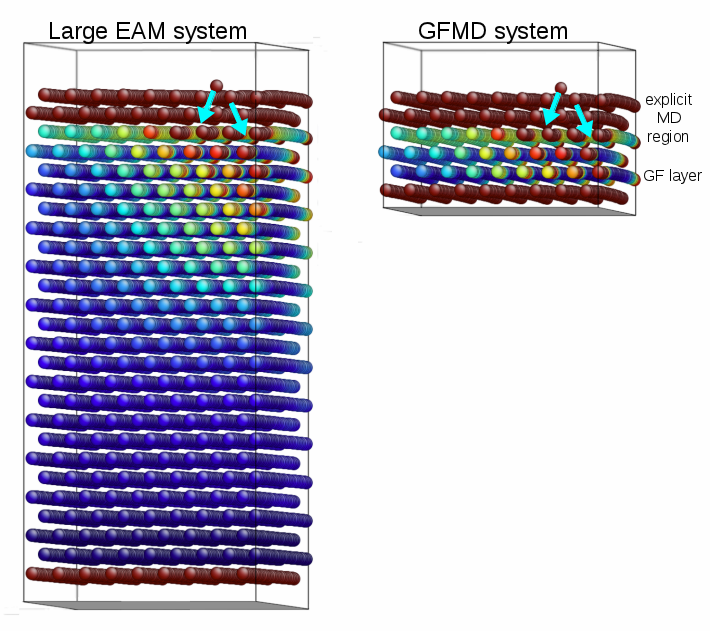
Friction between surfaces occurs at protrusions where the surfaces contact. These protrusions are often modeled as spherical bumps on the surface, and for this reason, the prototypical model of friction is a sphere sliding on a flat surface. Here, we use large simulations to find and explain new scaling of the friction force with asperity curvature [1].
On the atomic scale, the surfaces are composed of either crystalline or disordered material, and the materials may also have loose molecules adsorbed to their surfaces. In the simplest case, the two surfaces are crystalline and clean. Furthermore, it is simplest if the crystalline surfaces are aligned (ie commensurate) and if the surfaces do not stick together (ie are non-adhesive). The other situations are more complicated, though actually more common in every-day situations, and we analyze them in other work [2]. Here, we describe the friction in the simplest, cleanest case.
Most models of friction predict a friction coefficient, , with simple behavior. The Cattaneo-Mindlin model [3] assumes that each element of contacting surface area contributes a frictional force proportional to its normal force, with proportionality ,
. This leads to the standard friction behavior in introductory physics textbooks, where the friction force and the normal force being proportional to each other (Amonton’s Law):
(where
is the macroscopic friction coefficient and simple equals
). However, the atomic geometry leads to different behavior for these surfaces.
The effect of atomic geometry for this system had been considered before [4,5,6]. However, those previous studies used interactions characteristic of adhesive surfaces (tested in [2]), and did not simulate with atomic geometry. In our large-scale simulations of non-adhesive crystalline sphere sliding, we found unexpected behavior.
Fundamentally, to understand static friction, one must find the ways in which energy barriers arise in the system that oppose sliding. To understand kinetic friction, one must find the ways in which energy leaves the system. The energy is determined by the small scale atomic geometry at the surface and the long-range elastic interactions of the two materials.
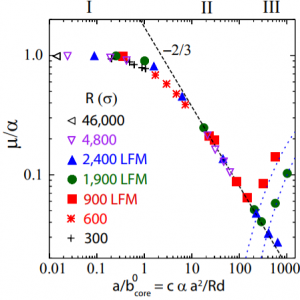
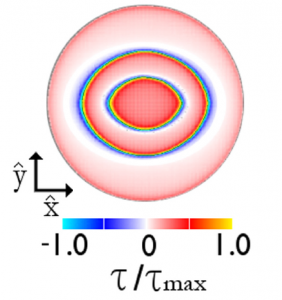
For the sphere sliding on a flat surface, elastic interactions distort the material during sliding in different ways depending on the amount of area in contact, leading to different values of the static friction coefficient. Under a sphere, the contact area is a circle, and is the contact radius.
is compared to a pressure-dependent material property,
, which is the core size of lattice dislocations at the interface between the two bodies. We found three distinct regimes of elastic distortions, for small, medium, and large contact radii, each producing a different emergent friction,
.
In regime I, the only elastic distortion comes from the sphere dragging the elastic substrate along the direction of motion. The stress, , builds at all places in the contact uniformly from
to the amount allowed by the local static friction
uniformly, where
is the local pressure. As in the Cattaneo-Mindlin model, the friction coefficient is equal to its microscopic value,
.
In regime II, stress builds primarily at the edges of the contact until a lattice dislocation develops and glides through the contact. The nucleation instability undercuts the ability of the friction to raise to , and the newly-uncovered scaling
can be derived. In regime III, dislocations are arrested in the contact, and so the friction comes from the Peierls stress to move dislocations. Surprisingly, the friction coefficient of a sphere of radius
rises with contact area, since the Peierls stress rises with pressure.
This surprising result of non-monotonic friction in the simplest manifestation of the sliding asperity model shows that friction still hides many behaviors waiting to be unraveled.
[1] Sharp, Tristan A. et al. “Scale-and load-dependent friction in commensurate sphere-on-flat contacts.” Physical Review B 96.15 (2017): 155436. https://journals.aps.org/prb/abstract/10.1103/PhysRevB.96.155436
[2] Sharp, Tristan A. et al. “Elasticity limits structural superlubricity in large contacts.” Physical Review B 93 (2016):121402(R) https://journals.aps.org/prb/abstract/10.1103/PhysRevB.93.121402
[3] Johnson, Kenneth. Contact mechanics. Cambridge university press, 1987.
Others’ work that helped build up this picture:
[4] Juan A. Hurtado and Kyung–Suk Kim. “Scale effects in friction of single–asperity contacts. I. From concurrent slip to single–dislocation–assisted slip.” Proceedings of the Royal Society A (1999): 455 1989 http://rspa.royalsocietypublishing.org/content/455/1989/3363.short
[5] Juan A. Hurtado and Kyung–Suk Kim. “Scale effects in friction of single–asperity contacts. II. Multiple–dislocation–cooperated slip” Proceedings of the Royal Society A (1999): 455 1989 http://rspa.royalsocietypublishing.org/content/455/1989/3363.short
[6] Gao, Yanfei. “A Peierls perspective on mechanisms of atomicfriction.” Journal of the Mechanics and Physics of Solids 58.12 (2010): 2023-2032. http://www.sciencedirect.com/science/article/pii/S0022509610001900